Genome-wide identification of bZIP transcription factors and their expression analysis in Platycodon grandiflorus under abiotic stress
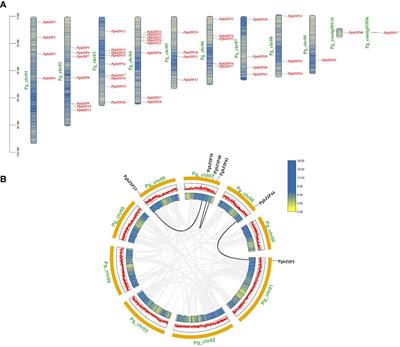
The Basic Leucine Zipper (bZIP) transcription factors (TFs) family is among of the largest and most diverse gene families found in plant species, and members of the bZIP TFs family perform important functions in plant developmental processes and stress response. To date, bZIP genes in Platycodon grandiflorus have not been characterized. In this work, a number of 47 PgbZIP genes were identified from the genome of P. grandiflorus, divided into 11 subfamilies.
The distribution of these PgbZIP genes on the chromosome and gene replication events were analyzed.
The motif, gene structure, cis-elements, and collinearity relationships of the PgbZIP genes were simultaneously analyzed. In addition, gene expression pattern analysis identified ten candidate genes involved in the developmental process of different tissue parts of P. grandiflorus. Among them, Four genes (PgbZIP5, PgbZIP21, PgbZIP25 and PgbZIP28) responded to drought and salt stress, which may have potential biological roles in P. grandiflorus development under salt and drought stress. Four hub genes (PgbZIP13, PgbZIP30, PgbZIP32 and PgbZIP45) mined in correlation network analysis, suggesting that these PgbZIP genes may form a regulatory network with other transcription factors to participate in regulating the growth and development of P.grandiflorus. This study provides new insights regarding the understanding of the comprehensive characterization of the PgbZIP TFs 3 for further exploration of the functions of growth and developmental regulation in P. grandiflorus and the mechanisms for coping with abiotic stress response.
Read the full article at the original website
References:
