Machine learning and weighted gene co-expression network analysis identify a three-gene signature to diagnose rheumatoid arthritis
Background: Rheumatoid arthritis (RA) is a systemic immune-related disease characterized by synovial inflammation and destruction of joint cartilage.
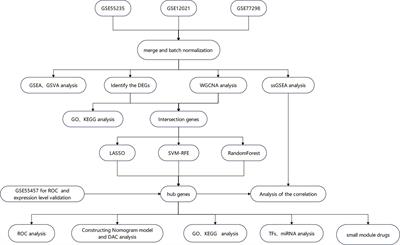
The pathogenesis of RA remains unclear, and diagnostic markers with high sensitivity and specificity are needed urgently. This study aims to identify potential biomarkers in the synovium for diagnosing RA and to investigate their association with immune infiltration. Methods: We downloaded four datasets containing 51 RA and 36 healthy synovium samples from the Gene Expression Omnibus database. Differentially expressed genes were identified using R.
Then, various enrichment analyses were conducted. Subsequently, weighted gene co-expression network analysis (WGCNA), random forest (RF), support vector machine–recursive feature elimination (SVM-RFE), and least absolute shrinkage and selection operator (LASSO) were used to identify the hub genes for RA diagnosis. Receiver operating characteristic curves and nomogram models were used to validate the specificity and sensitivity of hub genes. Additionally, we analyzed the infiltration levels of 28 immune cells in the expression profile and their relationship with the hub genes using single-sample gene set enrichment analysis. Results: Three hub genes, namely, ribonucleotide reductase regulatory subunit M2 (RRM2), DLG-associated protein 5 (DLGAP5), and kinesin family member 11 (KIF11), were identified through WGCNA, LASSO, SVM-RFE, and RF algorithms.
These hub genes correlated strongly with T cells, natural killer cells, and macrophage cells as indicated by immune cell infiltration analysis. Conclusion: RRM2, DLGAP5, and KIF11 could serve as potential diagnostic indicators and treatment targets for RA.
The infiltration of immune cells offers additional insights into the underlying mechanisms involved in the progression of RA.
Read the full article at the original website
References:
